Immobilization of modular peptides on graphene cocktail for differentiation of human mesenchymal stem cells to hepatic-like cells
- 1National Research Center for Transgenic Mouse, National Institute of Genetic Engineering and Biotechnology, Tehran, Iran
- 2Department of Clinical Biochemistry, Faculty of Medical Science, Tarbiat Modares University, Tehran, Iran
- 3Endocrinology and Metabolism Research Center, Endocrinology and Metabolism Clinical Sciences Institute, Tehran University of Medical Sciences, Tehran, Iran
- 4Department of Physics, Sharif University of Technology, Tehran, Iran
- 5Institute for Nanoscience and Nanotechnology, Sharif University of Technology, Tehran, Iran
- 6Nano Drug Delivery Research Center, Health Technology Institute, Kermanshah University of Medical Sciences, Kermanshah, Iran
- 7Department of Mycobacteriology and Pulmonary Research, Pasteur Institute of Iran, Tehran, Iran
- 8Dental Research Center, Hamadan University of Medical Sciences, Hamadan, Iran
- 9Dental Implant Research Center, Hamadan University of Medical Sciences, Hamadan, Iran
In this study, two novel biomimetic modular peptide motifs based on the alpha-2 subunit of type IV collagen (CO4A2) were designed and immobilized on a graphene platform to imitate integrin and heparan sulfate- (HS-) binding proteins. The in silico study was used to design 9-mer K[KGDRGD]AG and 10-mer KK[SGDRGD]AG for testing designed Integrin-Binding Peptide (dIBP) and HS-Binding Peptide (dHBP). The virtual docking technique was used to optimize the peptide motifs and their relevant receptors. Molecular dynamic (MD) simulation was used to evaluate the stability of peptide-receptor complexes. The effect of the platform on the differentiation of human mesenchymal stem cells (hMSCs) to hepatic-like cells (HLCs) was evaluated. After differentiation, some hepatic cells’ molecular markers such as albumin, AFP, CK-18, and CK-19 were successfully followed. Graphene-heparan sulfate binding peptide (G-HSBP) enhances the mature hepatic markers’ expression instead of control (p ≤ 0.05). The pathological study showed that the designed platform is safe, and no adverse effects were seen till 21 days after implantation.
Introduction
In advanced liver diseases, such as cirrhosis, many hepatocytes vanish, which cannot repair the liver tissue. Therefore, a liver transplant is required. However, because of the donor limitation and rejection problems, researchers urged to use cell therapy. In these patients, the lack of normal hepatocytes is a challenge. One choice is the differentiation of mesenchymal stem cells (MSCs) to hepatocyte-like cells (HLCs) on two-dimensional (2D) or three-dimensional (3D) biomimetic biomaterials (Jaiswal and Dhayal, 2020; Kraehenbuehl et al., 2011; Ramadas et al., 2017; Wang et al., 2020; Zujur et al., 2017). Using 2D or 3D microenvironments for differentiation purposes is preferable (Akhavan and Ghaderi, 2014; Lee et al., 2011; Suhito et al., 2017); however, it depends on the employed substrate bioactivation method (Asumda et al., 2018; Discher et al., 2009; Martino et al., 2009; Nayak et al., 2011; Kim et al., 2015; Stephanopoulos et al., 2015). Substrate bioactivation by ECM-derived proteins such as GAGs or HGF has disadvantages such as complex structural composition and immunogenic and labor working (Cai and Heilshorn, 2014; Graysona et al., 2009; Ghaedi et al., 2011; Lutolf and Hubbell, 2005). ECM-derived peptides as peptide motifs can mimic the function of ECM proteins with some benefits such as cheaper supplies, consistency, solubility, and exclusivity (Jhala and Vasita, 2015; Krishnamoorthy et al., 2018).
At this time, more than 250 peptide motifs are derived from ECM fibrous proteins. Most ECM-derived peptide motifs have been immobilized on different substrates, and their interactions with specific receptors have been studied. For example, GRGDSP, as a fibronectin-derived integrin binding peptide, activates α5β1 integrin (Defined three dimensional microenvironment for cell culture, 2018; Mota et al., 2013; Synthetically designed extracellular microenvironment, 2015). The most important issue with peptide immobilized biomaterials is their side effects and toxicity. Immobilized peptides cover the surface substrates preventing their interaction with cells and induce some essential mechanochemical signals for cell proliferation, development, and differentiation (Holle et al., 2008; Pan et al., 2016). Recently, MSC interaction with bioactive peptides immobilized on a graphene-based substrate is reported (Adibi-Motlagh et al., 2018). In this study, two novel biomimetic peptide motifs based on the alpha-2 subunit of type IV collagen (CO4A2) were designed and immobilized on a graphene platform to imitate integrin and heparan sulfate- (HS-) binding proteins for differentiation of MSCs to mature HLCs.
Materials and methods
All experimental protocols were approved by the Ethics and Animal Handling Committee of the Hamadan University of Medical Science (IR.UMSHA.REC.1397.357). All methods were conducted in accordance with relevant guidelines and regulations.
In silico study
6-mer peptide motif [GFPGER] and 7-mer peptide motif [TYRSRKY] were used as a positive control of Integrin-Binding Peptide (cIBP), and positive control of HS-Binding Peptide (cHBP), respectively (Hudalla and Murphy, 2010; Lee et al., 2007; Seo et al., 2010). Furthermore, two peptide motifs were selected from CO4A2, [KGDRGD] and [SGDRGD], which are used to design 9-mer K[KGDRGD]AG and 10-mer KK[SGDRGD]AG for testing Integrin-Binding Peptide (dIBP) and designed HS-Binding Peptide (dHBP).
Three-dimensional structure predictions
PEP-FOLD3 was used to predict the 3D structure of peptides (Lamiable et al., 2016). 3D structure of HS (ID Code:53477714) was downloaded from PubChem structure, and the crystal structure of αvβ3 Integrin (ID Code:1JV2) was downloaded from PDB.
Molecular docking study
Peptide motifs and their relevant receptors were optimized using Molegro Virtual Docker (MVD) version 4.0.2 (Thomsen and Christensen, 2006). Optimized structures were docked against peptides in 10 independent runs with the MolDock optimizer algorithm, and each run returned five poses. Poses obtained from 10 runs were ranked based on MolDock score, and the best score pose was selected as favorable peptide-receptor interaction. The simulation was repeated three times for each docking.
Molecular dynamic simulation study
A molecular dynamic (MD) simulation study was used to evaluate the stability of peptide-receptor complexes using GROMACS4.5.5 and GROMOSE 54A7 as force fields to generate proper topologies (Hess et al., 2008). The peptide-receptor complexes were put in a cubic box and filled with water using the TIP3P model. Cl− or Na+ ions were used as neutralizing agents. The steepest descent method was used for energy minimization. Periodic boundary conditions (PBC) were applied to avoid the edge effect interactions. All systems were equilibrated for 100 picoseconds (ps) under NVT and NPT at 300 K and 1 bar, respectively. 24,000 ps molecular dynamics simulation was run with no restraints. The root mean square plot (RMSD) shows the structural deviation of the peptide, which was plotted from the initial structure as a function of time. During the molecular dynamic simulation, peptide structure fluctuation was investigated by depicting root mean square fluctuation (RMSF). Binding free energy and the contribution of each residue in binding energy were calculated using the g_mmpbsa built-in function (available in GROMACS).
Peptide immobilization
Details of graphene oxide (GO) synthesis are reported elsewhere (Adibi-Motlagh et al., 2018). Graphene film was prepared by chemical reduction of a mold-derived GO. Briefly, GO solution (15 mg/ml) was added drop-by-drop to a plastic mold (plate) with a 5 cm radius. The fabricated graphene film was subsequently treated by chemical reduction with HI (40%) for 24 h at room temperature. The film was washed carefully with deionized (DI) water to remove the residual acid and iodine and freeze-dried to fabricate the graphene film. Peptide immobilization on the graphene film was made according to previous studies (Adibi-Motlagh et al., 2018; Rezaei et al., 2016). In a typical experiment, a methanol solution of cyclohexyl isocyanide, an aqueous solution of formaldehyde (1:1 eq), and peptide solution (1.1 eq) were added to the graphene film surface in MES buffer at pH 6.1. The system was shaken for 30 min. The film was subsequently washed with 0.01 M MES buffer. The obtained film was eluted with a 0.5 M NaCl solution and stored at 4 °C.
Cell culture, viability assay, and differentiation
5×105 human mesenchymal stem cells (hMSCs) were cultured in a modified culture dish at a 5% CO2 incubator (37°C). MTT assay was used to evaluate the cell viability of the platform. Briefly, 1 × 104 hMSCs were cultured in a 96-well plate, and after 24 and 48 h incubation times in a 5% CO2 incubator (37°C), the MTT solution was added to the wells. After 3 h of incubation, DMSO was added to the wells, and absorbance was read at 580 nm using an ELISA reader (Microplate reader labsystem multiscan). In order to differentiate hMSCs from HLCs, a two-step protocol was utilized. In the first step, early 7 days, cells were cultured in a 20 ng/ml HGF and 10–7 M DEX consisting of α-MEM supplemented with 10% FBS, and after cells attachment, in the 7th day, after the addition of inductive factors, 20 ng/ml HGF and 10–7 M DEX were followed for 2 weeks by 20 ng/ml OSM (Ghaedi et al., 2014).
RNA isolation and quantitative (q) real-time PCR
Total RNA was extracted from treated cells using a high pure RNA isolation kit (Roche, Germany). The extracted RNA was stored at −80°C until use. cDNA was synthesized by the Fermentas kit (Thermo Fisher, United States) and stored at −20°C until use. The real-time PCR was carried out with an ABI System (Applied Biosystems StepOne, United States) under the following thermal conditions: 95°C for 2°min, 40 cycles of 95°C for 10 s, and 58°C for 30 s. Melting curves were analyzed after 40 cycles to confirm the PCR product specificity. Primer sequences used for qReal-Time PCR analysis are presented in Supplementary Table S1.
Animal study
Fourteen-week adult Wistar rats were housed in controlled temperature and humidity conditions (20 ± 2°C on a 12:12 h light/dark cycle). Animals were shaved following anesthetization by ketamine/xylazine (100/12.5 mg/kg) procedure. The sterile designed platform was implanted subcutaneously between skin and fascia. The animal was closed by surgical suture and followed up for 3 weeks.
Statistical analysis
All experiments were carried out in triplicate. The Shapiro–Wilk method was used to evaluate the data distribution. Student’s t-test and analysis of variance (ANOVA) were used to analyze experimental data. SPSS 21 software was utilized for analysis. p < 0.05 was considered statistically significant.
Results and discussion
In silico study
Virtual docking
Docking designed peptides with αvβ3 integrin and HS receptor
In this study, two novel CO4A2-derived peptide motifs, KGDRGD and SGDRGD, were designed. For this purpose, different peptides were docked against HS with three repeated runs. As shown in Figure 1A, the highest docking score is related to the peptides-integrin complex. However, dIBP and dHBP did not show a significant difference in binding to the receptor, and peptides exhibit the same behavior as cIBP-integrin docking. For adapting to a high affinity-repeat sequence and more flexibility, peptide motifs were modified to KK[SGDRGD]AG and K[KGDRGD]AG, which were designed as a ligand with high-affinity binding to HS, designed HS binding motif (dHBP) and binding to integrin receptor, designed integrin binding peptide motif (dIBP), respectively. Figure 1B shows the highest docking score of the peptides-HS complex. The control peptide motif (cHBP) binding to HS had no significant difference with the designed peptide (dHBP). Despite the similarity of high sequence to peptide, the peptide motif interaction with integrin receptor (dIBP) shows a lower affinity to HS (Figure 2).
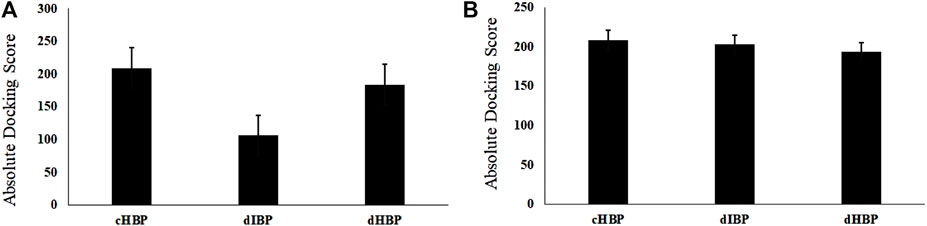
FIGURE 1. Graph of (A) integrin docking score; (B) HS docking score with designed peptides compared to positive control.
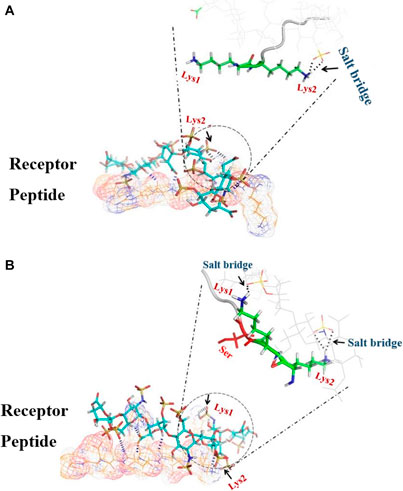
FIGURE 2. 3D image of the dP-HS complex; the main interaction energy comes from hydrogen bonds (blue dashes) and salt bridges (black dashes). (A) In dIBP-HS, only Lys2 forms a salt bridge (black arrow) with HS. (B) In dHBP-HS, both Lys1 and Lys2 form a salt bridge with HS.
Meanwhile, our bioinformatics studies complied with the results of Imberty et al., suggesting that cytokines through repeat sequence (KKXXXK) can bind to HS, but another repeat sequence (KKXXK) has less affinity to HS (Lortat-Jacob et al., 2002). A molecular docking study showed that the highest docking score with HS is related to cHSBP and followed by dHBP. Score docking of dIBP, despite the similarity to dHBP, shows the least susceptibility to HS. In fact, the presence of the third residue in Serine amino acid (3S) in the vicinity of lysine amino acid (2K) is a spatial position to form a salt bridge with HS (Figure 2A). In contrast, in the absence of residue 3S in dIBP, the Amine Lysine # 2 acid is not in a suitable position and cannot form a salt bridge with HS (Figure 2B). In fact, the presence of 3S in the vicinity of 2K approximated this residue to HS and salt bridge formation.
Molecular dynamics simulation
Supplementary Figure S1 shows the RMSD of carbon alpha Cα for two peptides in complex with HS during simulation. Compared to dIBP, dHBP indicates lower RMSD, especially at the last frames (0.36 vs. 0.55). Therefore, Cα backbone peptide atoms in the dHBP-HS complex have a less deviation than the dIBP-HS complex over the last 24,000 ps. Therefore, there is no significant difference between docking results of designed peptides with integrin.
As shown in the plot, Cα atoms of dHBP peptides in complex with HS have less fluctuation during the simulation than dIBP, indicating more stability of the dHBP-HS complex (Supplementary Figure S1). Moreover, HS atoms are more rigid and stable in binding to dHBS (Supplementary Figure S1). In contrast, the total energy of dHBP-HS (blue) is twofold more than dIBP-HS (red in Supplementary Figure S1).
Cell morphology and viability
Morphological study of differentiated hepatic-like cells on the modified substrates shows that on the 7th day after the culture, cell differentiation was initiated. However, the microscopy images revealed that these changes were remarkable and clear on the 14th day after the culture; the differentiated cells were spherical and tended to make a clear colony (Figure 3B). After 21 days, the shape and morphology of cells were semi-HLCs. Cellular viability on G-dHBP, G-dIBP, and graphene platforms, after 24 and 48 h, were 96%, 97% and 93%, 92%, and 90% and 87%, respectively.
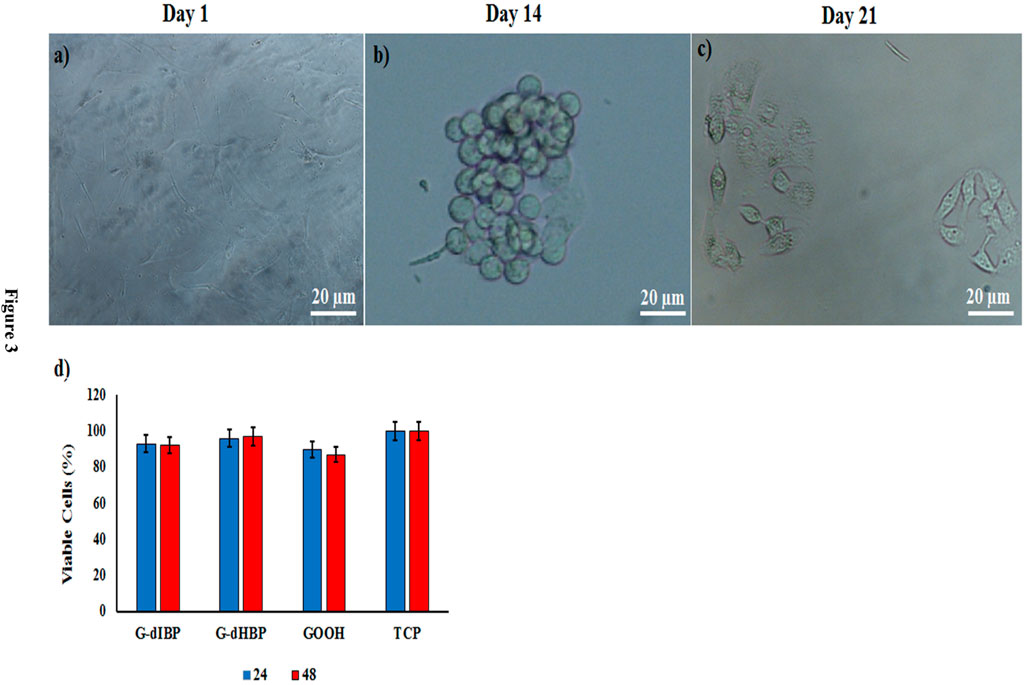
FIGURE 3. Time-dependent kinetics of MSCs differentiation to HLCs (A,B,C). (D) Viability of cultured cells on different platforms. TCP indicates Tissue Culture Plate as control.
CK18 and CK19 quantification
The CK18 and CK19 transcript levels were studied as the main hepatic cytoskeleton proteins. The obtained results showed that, on the 14th day after differentiation initiation, the CK18 transcript level was ∼1.4 for all groups, but it was greater in the G-dHBP platform (Figure 4A). As long as the differentiation time was 21 days after differentiation initiation, the transcript level reached 2.5-fold in the G-dHBP platform, which was statistically significant. However, the CK19 transcript level had no significant difference in different groups during the differentiation and even on the 21st day after differentiation initiation (Figure 4B). The mean quantitative gene expression of liver cytoskeleton proteins is presented in Figure 4. Comparison between CK18 and CK19 on the substrate containing dIBP, dHBP peptide, and controls was investigated on the 14 and 21 days after the cell culture (Figure 4). The mean CK18/CK19 ratio in differentiated cells on G-dHBP was higher than the rest of the groups, and on the 21st day after the culture, it was 2.5-fold. Furthermore, in the G-dIBP group, this ratio was greater than in controls (GOOH and TCP) (Figure 4C).
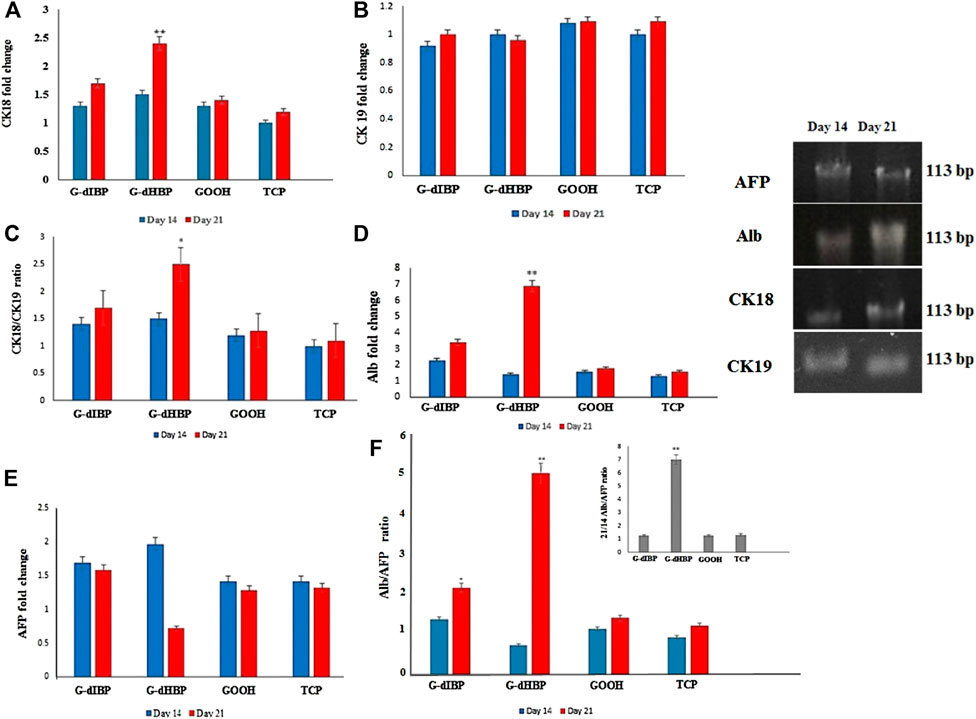
FIGURE 4. qRT-PCR of (A) CK18, (B) CK19, (C) CK18/CK19, (D) Alb, (E) AFP, and (F) Alb/AFP expression in different groups at 14 and 21 days after differentiation.
As the differentiation process has time-dependent kinetics, comparing the mature to immature marker ratios approves the process and validates the molecular dynamic simulation. Alb is a dominant protein produced by hepatocytes, which begins with initial embryonic hepatocytes and reaches the maximum in adult hepatocytes. AFP is a “fetal initial” marker, and its expression is reduced by liver development. Alb expression is an important marker of mature hepatocytes on the 21st day of differentiation, whereas AFP is a marker for immature hepatocytes. Fourteen days after the culture, the cell differentiation peaked, and AFP expression as an embryonic cell functional protein (immature) was turned-off instead of the functional protein of hepatocyte cells.
Albumin and alpha-fetoprotein quantification
Quantitative real time-PCR was applied to obtain the marker genes transcript level in differentiated cells. Alpha-fetoprotein (AFP) is an embryonic factor mainly expressed in the early stage of cell development as long as cells mature. Quantitative real-time PCR results showed that the AFP transcript level was significantly decreased in the G-dHBP platform 21 days after the differentiation (p ≤ 0.05) (Figure 4E).
In order to confirm the specificity effect of G-dHBP on hepatocyte-stage (maturity stage of HLCs), the cytoskeleton rearrangement markers expression (CK18/CK19) ratios were obtained (Figure 4C). Albumin (Alb) is a mature hepatic cell protein marker. The results of this study show that the Alb transcript level in differentiated cells in the G-dHBP platform (21 days after differentiation) was significantly greater (sevenfold) than that in other groups (p ≤ 0.01) (Figure 4D). The Alb/AFP expression ratio is obtained in all platforms. The results of this study show that the mean Alb/AFP expression ratio in differentiated cells at 14 and 21 days after differentiation is as follows: after 21 days of differentiation, the G-dHBP platform was significantly greater in all groups (p ≤ 0.001) (red dHBP column in Figure 4F). The ratio of 14 days after differentiation in the G-dHBP substrate was not significantly greater than that in other groups (Figure 4F). The Alb/AFP ratio was monitored at 14–21 days after differentiation (Figure 4F). The results show that at 21 days after differentiation, the Alb/AFP ratio was significantly greater in the G-dHBP platform (sevenfold) (p ≤ 0.001). However, the Alb/AFP ratio was not significantly different between G-dIBP, GOOH, and TCP (Figure 4F). The comparison between the ratio of HLCs mature to fetal marker (Alb/AFP) at 14 and 21 days after differentiation shows that this ratio reached its peak on day 21, whereas it was reversed on day 14 (Figure 4F). Briefly, the 21/14 Alb/AFP ratio increased sevenfold compared to controls (G-dIBP and GOOH) and TCP (Figure 4F).
Platform
The graphene immobilized peptide synthesis and characterization were reported elsewhere (Adibi-Motlagh et al., 2018; Rezaei et al., 2016).
HMSC cells attached to the platform are shown in Figure 5E. As shown in Figure 5F, after 14 days of onset differentiation on the platform, the semi-hepatic phenotype cells were formed. Rolling up cells and their multi-face mode seems to be time-dependent. Cell morphological change is presented in Figures 5E,F. As shown in Figure 3, the mature cell nucleus and cytoplasm are rich in small vacuoles.
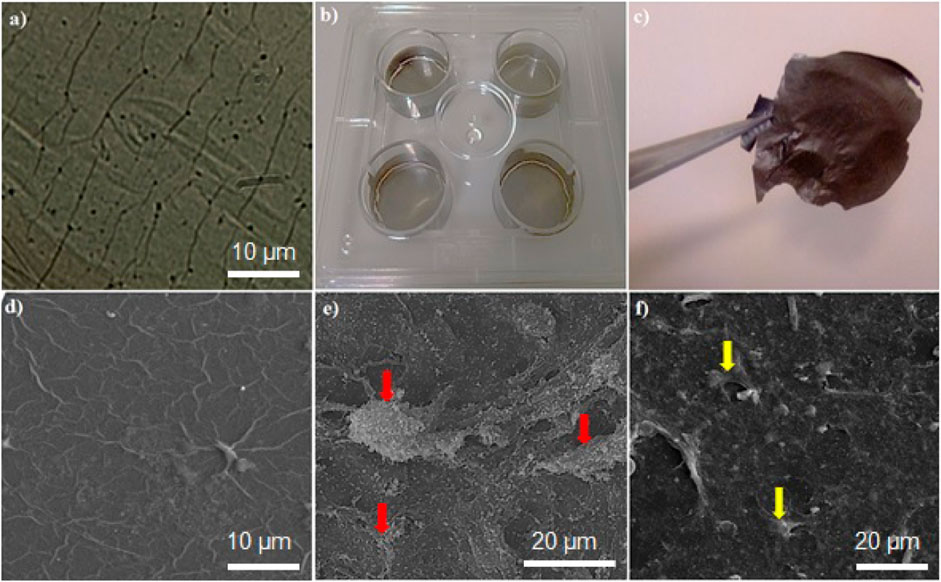
FIGURE 5. (A) Microscopic image of the graphene-based platform, (B) plate coated by graphene platform, (C) graphene film; SEM images of (D) the graphene-based platform, (E) hMSCs attached on graphene platform, (F) differentiated cells on the graphene-based platform. Red and yellow arrows show hMSCs and differentiated cells, respectively.
Histopathology
After 3 weeks of implantation of the graphene-based platform, the platform implanted in the livers and spleens of rats was checked (Figures 6A–C) and rats were followed up during this time period. During this time, no adverse effects, including feeding and drinking, or changes in animal weights during the treatment were observed. Histopathological results revealed no structural change in liver cells of treated rats with the implanted graphene-based platform. Veins, Kupfer, and hepatocyte cells were normal, and no remarkable differences between treated and non-treated samples were seen (Figures 6D,E). Normal architecture in both treated and control groups was clearly seen (Figures 6F,G, normal white and red pulps with any damages). The observation data and histopathological results revealed that the synthesized platforms are biocompatible in the in vivo system and have no side effects when it comes to in vivo applications.
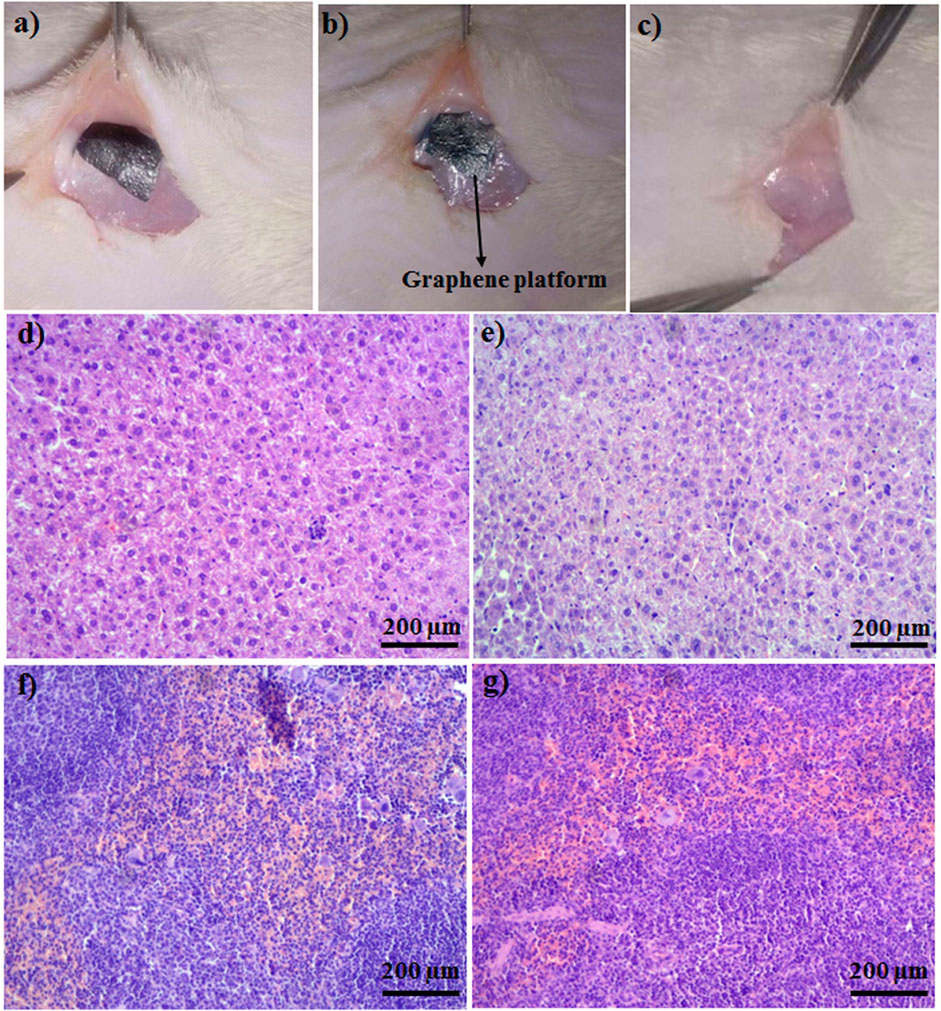
FIGURE 6. (A) Platform was implanted subcutaneously, (B) platform after 2 days of implantation, (C) control. Pathology images of (D) liver of control, (E) platform implanted liver, (F) spleen of control, (G) platform implanted spleen.
Conclusion
In conclusion, graphene platform and peptide motifs as physical and biochemical factors were used to make a biological cocktail for the regulation of in vitro cell fate. The biomimetic cocktail differentiated hMSCs to mature HLCs. The maturity of generated cells is mainly due to the use of designed peptide motifs that naturally manipulate the cell behavior. By applying the in silico approach, functional peptides were selected that, by making a biomimetic cocktail, successfully generated mature hepatic cells.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by the Ethics committee of the Hamadan University of Medical Science.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Acknowledgments
The authors would like to thank the Research Council Hamedan University of Medical Sciences (9706203608) for their financial support and provision of facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.943003/full#supplementary-material
References
Adibi-Motlagh, B., Sahebghadam Lotfi, A., Rezaei, A., and Hashemi, E. (2018). Cell attachment evaluation of the immobilized bioactive peptide on a nanographene oxide composite. Mater Sci. Eng. C 82, 323–329. doi:10.1016/j.msec.2017.05.039
Akhavan, O., and Ghaderi, E. (2014). The use of graphene in the self-organized differentiation of human neural stem cells into neurons under pulsed laser stimulation. J. Mat. Chem. B 2, 5602–5611. doi:10.1039/c4tb00668b
Asumda, F. Z., Hatzistergos, K. E., Dykxhoorn, D. M., Jakubski, E. D. M., and Edwards, E. R. (2018). Differentiation of hepatocyte-like cells from human pluripotent stem cells using small molecules. Differentiation 101, 16–24. doi:10.1016/j.diff.2018.03.002
Cai, L., and Heilshorn, S. C. (2014). Designing ECM-mimetic materials using protein engineering. Acta Biomater. 10, 1751–1760. doi:10.1016/j.actbio.2013.12.028
Discher, D. E., Mooney, D. J., and Zandstra, P. W. (2009). Growth factors, matrices, and forces combine and control stem cells. Science 324, 1673–1677. doi:10.1126/science.1171643
Ghaedi, M., Duan, Y., Zern, M. A., and Revzin, A. (2014). Hepatic differentiation of human embryonic stem cells on growth factor-containing surfaces. J. Tissue Eng. Regen. Med. 8, 886–895. doi:10.1002/term.1595
Ghaedi, M., Tuleuova, N., Zern, M. A., Wu, J., and Revzin, A. (2011). Bottom-up signaling from HGF-containing surfaces promotes hepatic differentiation of mesenchymal stem cells. Biochem. Biophys. Res. Commun. 407, 295–300. doi:10.1016/j.bbrc.2011.03.005
Graysona, W. L., Martensa, T. P., and Eng, G. M. (2009). Biomimetic approach to tissue engineering. Stem Cell. Dev. Biol. 20, 665–673. doi:10.1016/j.semcdb.2008.12.008
Hess, B., Kutzner, C., van der Spoel, D., and Lindahl, E. (2008). Gromacs 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447. doi:10.1021/ct700301q
Holle, A. W., Tang, X., Vijayraghavan, D., Vincent, L. G., Fuhrmann, A., Choi, Y. S., et al. (2008). In situ mechanotransduction via vinculin regulates stem cell differentiation. Stem cells Dev. 17, 29–36.
Hudalla, G. A., and Murphy, W. L. (2010). Immobilization of peptides with distinct biological activities onto stem cell culture substrates using orthogonal chemistries. Langumir 26, 6449–6456. doi:10.1021/la1008208
Jaiswal, J., and Dhayal, M. (2020). Preparation of 2D coatings of functionally graded chitosan-gold nanocomposite through in-situ reduction in cationic and anionic environments: Application for inhibiting hepatocellular carcinoma cells response. Mater. Chem. Phys. 243, 122663. doi:10.1016/j.matchemphys.2020.122663
Jhala, D., and Vasita, R. (2015). A review on extracellular matrix mimicking strategies for an artificial stem cell niche. Polym. Rev. 55, 561–595. doi:10.1080/15583724.2015.1040552
Kim, T. H., Shah, S., Yang, L., Yin, P. T., Hossain, M. K., Conley, B., et al. (2015). Controlling differentiation of adipose-derived stem cells using combinatorial graphene hybrid-pattern arrays. ACS Nano 9, 3780–3790. doi:10.1021/nn5066028
Kraehenbuehl, P., Langer, L. R., and Ferreira, M. (2011). Three-dimensional biomaterials for the study of human pluripotent stem cells. Nat. Methods 8, 731–736. doi:10.1038/nmeth.1671
Krishnamoorthy, N., Tseng, Y. T., Gajendrarao, P., Sarathchandra, P., McCormack, A., Carubelli, I., et al. (2018). A strategy to enhance secretion of extracellular matrix components by stem cells: Relevance to tissue engineering. Tissue Eng. Part a. 24, 145–156. doi:10.1089/ten.tea.2017.0060
Lamiable, A., Thévenet, P., Rey, J., Vavrusa, M., Derreumaux, P., and Tufféry, P. (2016). PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 44, W449–W454. doi:10.1093/nar/gkw329
Lee, J. Y., Choo, J. E., Choi, Y. S., Lee, K. Y., Min, D. S., Pi, S. H., et al. (2007). Characterization of the surface immobilized synthetic heparin binding domain derived from human fibroblast growth factor-2 and its effect on osteoblast differentiation. J. Biomed. Mater Res. A 83, 970–979. doi:10.1002/jbm.a.31351
Lee, W. C., Lim, C. H., Shi, H., Tang, L. A., Wang, Y., Lim, C. T., et al. (2011). Origin of enhanced stem cell growth and differentiation on graphene and graphene oxide. ACS Nano 5, 7334–7341. doi:10.1021/nn202190c
Lortat-Jacob, H., Grosdidier, A., and Imberty, A. (2002). Structural diversity of heparan sulfate binding domains in chemokines. Proc. Natl. Acad. Sci. U. S. A. 99, 1229–1234. doi:10.1073/pnas.032497699
Lutolf, M. P., and Hubbell, J. A. (2005). Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 23, 47–55. doi:10.1038/nbt1055
Martino, M. M., Mochizuki, M., Rothenfluh, D. A., Rempel, S. A., Hubbell, J. A., and Barker, T. H. (2009). Controlling integrin specificity and stem cell differentiation in 2D and 3D environments through regulation of fibronectin domain stability. Biomaterials 30, 1089–1097. doi:10.1016/j.biomaterials.2008.10.047
Mota, A., Sahebghadam Lotfi, A., Barzin, J., Hatam, M., Adibi-Motlagh, B., Khalaj, Z., et al. (2013). Human bone marrow mesenchymal stem cell behaviors on PCL/gelatin nanofibrous scaffolds modified with a collagen IV-derived RGD-containing peptide. Cell. J. (YAKHTEH) 15, 1–27.
Nayak, T. R., Andersen, H., Makam, V. S., Khaw, C., Bae, S., Xu, X., et al. (2011). Graphene for controlled and accelerated osteogenic differentiation of human mesenchymal stem cells. ACS Nano 5, 4670–4678. doi:10.1021/nn200500h
Pan, G., Sun, S., Zhang, W., Zhao, R., Cui, W., He, F., et al. (2016). Biomimetic design of mussel-derived bioactive peptides for dual-functionalization of titanium-based biomaterials. J. Am. Chem. Soc. 138, 15078–15086. doi:10.1021/jacs.6b09770
Ramadas, M., Bharath, G., Ponpandian, N., and Ballamurugan, A. M. (2017). Investigation on biophysical properties of Hydroxyapatite/Graphene oxide (HAp/GO) based binary nanocomposite for biomedical applications. Mater. Chem. Phys. 199, 179–184. doi:10.1016/j.matchemphys.2017.07.001
Rezaei, A., Akhavan, O., Hashemi, E., and Shamsara, M. (2016). Ugi four-component assembly process: An efficient approach for one-pot multifunctionalization of nanographene oxide in water and its application in lipase immobilization. Chem. Mat. 28 (2016), 3004–3016. doi:10.1021/acs.chemmater.6b00099
Thomsen, R., and Christensen, M. H. (2006). MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 49, 3315–3321. doi:10.1021/jm060961+
Seo, N., Russell, B. H., Rivera, J. J., Liang, X., Xu, X., Afshar-Kharghan, V., et al. (2010). An engineered integrin-binding collagenous sequence. J. Biol. Chem. 285 (2010), 31046–31054. doi:10.1074/jbc.m110.151357
Stephanopoulos, N., Freeman, R., North, H. A., Sur, S., Jeong, S. J., Tantakitti, F., et al. (2015). Bioactive DNA-peptide nanotubes enhance the differentiation of neural stem cells into neurons. Nano Lett. 15, 603–609. doi:10.1021/nl504079q
Suhito, I. R., Han, Y., Kim, Y., Son, H., and Kim, T. H. (2017). Effects of two-dimensional materials on human mesenchymal stem cell behaviors. Biochem. Biophysical Res. Commun. 493, 578–584. doi:10.1016/j.bbrc.2017.08.149
Wang, J., Zhang, Y., Yang, X., and Ma, X. (2020). Stress effect on 3D culturing of MC3T3-E1 cells on microporous bovine bone slices. Nanotechnol. Rev. 9, 1315–1325. doi:10.1515/ntrev-2020-0103
Keywords: graphene platform, differentiation, hepatic-like cells, in silico, modular peptide
Citation: Adibi-Motlagh B, Hashemi E, Akhavan O, Khezri J, Rezaei A, Zamani Amir Zakria J, Siadat SD, Sahebghadam Lotfi A and Farmany A (2022) Immobilization of modular peptides on graphene cocktail for differentiation of human mesenchymal stem cells to hepatic-like cells. Front. Chem. 10:943003. doi: 10.3389/fchem.2022.943003
Received: 13 May 2022; Accepted: 29 July 2022;
Published: 29 August 2022.
Edited by:
Daniela Caschera, National Research Council (CNR), ItalyReviewed by:
Andrei I. Khlebnikov, Tomsk Polytechnic University, RussiaUttam Pal, Indian Institute of Chemical Biology (CSIR), India
Copyright © 2022 Adibi-Motlagh, Hashemi, Akhavan, Khezri, Rezaei, Zamani Amir Zakria, Siadat, Sahebghadam Lotfi and Farmany. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abbas Sahebghadam Lotfi, lotfi_ab@modares.ac.ir; Abbas Farmany, a.farmany@ut.ac.ir
 Behzad Adibi-Motlagh1,2
Behzad Adibi-Motlagh1,2  Ehsan Hashemi
Ehsan Hashemi Omid Akhavan
Omid Akhavan Abbas Sahebghadam Lotfi
Abbas Sahebghadam Lotfi Abbas Farmany
Abbas Farmany